October 2016 Galaxy News
- Galaxy Australia Community Launched
- Who's Hiring
- Trinity comes to usegalaxy.org
- Galaxy Community Hubs
- ToolShed Contributions
- Other News
Welcome to the October 2016 Galactic News, a summary of what is going on in the Galaxy community.
The big news this month is a series of upcoming events:
-
Galaxy training contribution fest:
- October 6-7, Online
-
- October 20-21, Freiburg, Germany
-
- Month of October, online
-
- November 2-3, Online
-
- November 7-11, Salt Lake City, Utah
- On time registration ends October 16
-
Galaxy Australasia Meeting (GAMe 2017):
- February 3-9, Melbourne, Victoria, Australia
- Registration and abstract submission are now open.
If you have anything to include in the next News, please send it to [Galaxy Outreach](mailto:outreach AT galaxyproject DOT org).
Events
There are a wealth of events coming up. Here are some highlights, and a report on one from last month too.
Galaxy Australasia Meeting (GAMe 2017): Registration & Abstract Submission are Open
We are pleased to announce that early registration and talk and poster abstract submission are now open for GAMe 2017. This meeting will bring together biomedical researchers, bioinformaticians, infrastructure providers, and data producers from across Australia and Asia to share expertise across many levels.
The conference starts 4 February and features two full days of keynotes, accepted and sponsor talks, poster and sponsor sessions, birds-of-a-feather gatherings, a conference dinner and lots of opportunities for networking.
Prior to the conference we are offering a Researcher Training Day on 3 February, aimed at biomedical researchers who need to analyse their biological data. The conference is followed by a four day workshop on Galaxy server administration for those working at that level.
Talk abstracts are due 30 November and early registration ends 31 December. Register now and save up to 43% off regular registration rates. There are also scholarships available for eligible students and postdocs. The scholarship application deadline is 18 November.
We look forward to welcoming you to Melbourne. Remember to pack for our fabulous February weather!
GAMe 2017 Organising Committee (#game_2017)
And if you are based in Australia you might also be interested in the newly launched Galaxy Australia Community.
Galaxy training contribution fest, 6-7 October, Online
The Galaxy Training Network (GTN) is planning a contribution fest to consolidate our training material and improve the overall training experience in Galaxy on the 6.-7th October 2016!
More information about the project can be found here and a small presentation here. Registration (which is entirely optional - you can just show up) can be done via this GitHub issue. There is also a list of potential issues to work on as well (and more are welcome).
This will be an online contribution fest and the fun to contribute should be priority one! There is enough to work on in all areas, from programming to fix spelling mistakes. So everyone is welcome to improve the Galaxy training material.
Ciao,
Galaxy Admin Training, November 7-11, Salt Lake City, Utah

November 7-11, 2016
University of Utah
Salt Lake City, Utah
United States
[Register by October 16](/events/admin-training2016/registration/)
Register for Galaxy Admin Training 2016 by October 16.
Galaxy Admin Training 2016 is a weeklong event offering basic and advanced sessions. It is being held in Salt Lake City, Utah, November 7-11, 2016, the week before Supercomputing 16 (SC16) meets in Salt Lake.
This two day basics session will introduce participants to what you need to know to get a Galaxy server up and running on a standalone server. You'll also learn how to extend your Galaxy with your own tools and tools from the community, and how to define reference data in your server as well.
Advanced Session: November 9-11
The three day advanced session will build on topics covered in the basics section. This session both extends those topics, such as tool definition, and adds new ones, such as working with compute clusters and heterogeneous resources. The goal of the advanced workshop is to enable Galaxy administrators to create robust, high-performance Galaxy instances that take full advantage of available tools and compute and storage resources.
Register by October 16 and save up to $230 off late registration rates (and up to $500 if you work in industry). Space is also limited.
Galaxy Admin Training 2016 is hosted by the University of Utah Center for High Performance Computing (CHPC), the USTAR Center for Genetic Discovery (UCGD), the Department of Biomedical Informatics and the Clinical & Translational Science Biomedical Informatics Core (CCTS BMIC) at the University of Utah.
We hope to see you in Salt Lake!
Swiss-German Galaxy Days
The 2016 editions of Swiss-German Galaxy Days will be held 20-21 October in Freiburg (Germany).
Registration is free, but space is limited on both days. We recommend you register soon to secure your spot for one or both days of the SG2016Tour:
The two days will be held in the same location, but will have a different focus. The first day will start with talks, discussions and small demonstrations. This day will be problem / technology centric with small demonstrations and hands-on sessions. Everyone is expected to have their own computer.
The second day is a developers day and we are planning to have 3 to 4 hands-on sessions (1.5h). Every session will start with a 10 minute talk and theoretical introduction, followed by 1h of hacking and 20 minutes discussion and a small break until the next session begins. We have a few proposed and prepared topics but we are flexible so we can discover new areas of the Galaxy.
Galaxy & Hacktoberfest
Galaxy and CloudBridge have joined the Hacktoberfest event this year. If you register with your GitHub account and create 4 or more pull requests during October you will get a free T-shirt and world-wide appreciation! You can browse the entry level issues in the Galaxy code by checking the hacktoberfest label (Galaxy, CloudBridge). CloudBridge already has a new contributor so hurry and hack away!
Conda, Conda, Conda!
The Galaxy v16.07 release (see below) included a "Shift of Galaxy tool dependencies to Conda" and that shift is reflected in a whole bunch of activity:
- Conda as a new standard for Galaxy tool dependencies, a guest blog posting by Björn Grüning in GigaScience's GigaBlog.
- a Galaxy-P Conda contribution fest in September (see report below),
- enhancements to Planemo, and
- an upcoming Bioconda Contribution Fest on November 2-3.
Galaxy-P Conda Contribution Fest Report
The Galaxy-P Conda Contribution Fest was held online, 27-28 September, with the aim to port as many tool dependencies to Conda packages as possible. Conda packages are the future of Galaxy’s tool dependency management and will yield more robust tools and automatic CI testing of all Galaxy-P tools. More than 20 pull requests were create and merged to BioConda. See the event report for details.
Thanks to everyone who participated!
All upcoming events
There are a plentitude of Galaxy related events coming up in the next few months:
| |
Designates a training event offered by GTN member(s) |
See the Galaxy Events Google Calendar for details on other events of interest to the community.
Galaxy Australia Community Launched
Today we officially launch the Galaxy Australia community with the opening of registrations for the Galaxy Australasia Meeting 2017 (GAMe 2017) being held in Melbourne over 3-9 February. Australian Galaxy users and administrators will now have more opportunities to interact and collaborate to identify and address the needs of the local community. The community will also be a hub for Australian Galaxy training courses and offer a catalog of Australian Galaxy servers.
The establishment of Galaxy Australia is supported by the EMBL Australia Bioinformatics Resource (EMBL-ABR) in its appointed role of supporting the development of skills and training in the Australian life science research community. At the EMBL-ABR Hub, hosted at the Victorian Life Sciences Computation Initiative (VLSCI), Galaxy is used extensively for teaching and data analysis directly as well as through the Australian-made Genomics Virtual Laboratory. VLSCI’s expert bioinformaticians and professional staff will help promote community activities and training.
Galaxy Australia has a has a Twitter feed (@GalaxyAustralia) for announcements and discussion relevant to the community.
Galaxy Australasia Meeting 2017 (GAMe 2017)
Our first activity is to organise the Galaxy Australasia Meeting 2017 (GAMe 2017), 3-9 February in Melbourne. Registration and abstract submission are open. See the announcement for full details.
New Publications
117 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in September. The total number of publication is now over 3600.
Some highlights from last month:
The Essential Components of a Successful Galaxy Service Annette McGrath, Steve McMahon, Sean Li, Joel Ludbey, Tim Ho. Journal of Grid Computing (2016), pp. 1-11, doi:10.1007/s10723-016-9379-6
Enhancing pre-defined workflows with ad hoc analytics using Galaxy, Docker and Jupyter Björn Grüning, Helena Rasche, Boris Rebolledo-Jaramillo, Carl Eberhard, Torsten Houwaart, John Chilton, Nathan Coraor, Rolf Backofen, James Taylor, Anton Nekrutenko. bioRxiv (16 September 2016), 075457, doi:10.1101/075457
Genotyping of evolving prokaryotic populations Markus Zojer, Lisa N. Schuster, Frederik Schulz, Alexander Pfundner, Matthias Horn, Thomas Rattei. PeerJ Preprints (14 September 2016), doi:10.7287/peerj.preprints.2449v1
Identifying Regions Enriched in a ChIP-seq Data Set (Peak Finding) Jui-Hung Hung, Zhiping Weng. Cold Spring Harbor Protocols (29 August 2016), doi:10.1101/pdb.prot093187
Metavisitor, a suite of Galaxy tools for simple and rapid detection and discovery of viruses in deep sequence data Guillaume Carissimo, Marius van den Beek, Juliana Pegoraro, Kenneth D. Vernick, Christophe Antoniewski. bioRxiv (2016), doi:10.1101/048983
Cloudflow - A framework for MapReduce pipeline development in Biomedical Research Lukas Forer, Enis Afgan, Hansi Weißensteiner, Davor Davidović, Günther Specht, Florian Kronenberg, Sebastian Schönherr. 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) (May 2015), pp. 172-177, doi:10.1109/mipro.2015.7160259
Tagged Publications
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | Cloud | 5 | Other | 3 | Shared | 9 | UseMain | |||
| 4 | HowTo | 3 | Project | 6 | Tools | 27 | UsePublic | |||
| 2 | IsGalaxy | 4 | RefPublic | - | UseCloud | 1 | Visualization | |||
| 58 | Methods | 9 | Reproducibility | 9 | UseLocal | 31 | Workbench |
Who's Hiring

The Galaxy is expanding! Please help it grow.
- 6 month position at Public Health England working National Microbiology Laboratory in Canada to compare antimicrobial resistance isolates and software using IRIDA/Galaxy
- Intégration d'outils pour la métagénomique phytovirale dans Galaxy, INRA, Bordeaux, France
- Postdoctoral Research Position, Multiomic Bioinformatics, University of Minnesota
- Postdoctoral Researcher: Forest Genomics Database and Software Developer, University of Connecticut. "... data sharing among partner databases through Galaxy modules." Act fast.
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
Public Galaxy Server News
There are over 80 publicly accessible Galaxy servers and five semi-public Galaxy services. Here's what happened with them in September.
New Public Galaxy Servers
Galaxy-CEFAP
Galaxy-CEFAP offers a set of tools to perform RNA-Seq and miRNA. User support is available via [Email](mailto:geninfo AT icb DOT usp DOT br), and there is an FTP tutorial as well. Anyone can use the server, but it is necessary to create a login first. Email [geninfo AT icb DOT usp DOT br](mailto:geninfo AT icb DOT usp DOT br) to request an account. There is a storage quota for all users.
Galaxy-CEFAP is supported by Instituto de Ciências Biomédicas - ICB, Universidade de São Paulo - USP, and Fundação de Amparo à Pesquisa de SP - FAPESP hosted by Centro de Facilidades de Apoio à Pesquisa - CEFAP-USP.
VarCap
VarCap Galaxy performs reliable prediction of different types of variants even at low frequencies. See Zojer M, Schuster LN, Schulz F, Pfundner A, Horn M, Rattei T. (2016) Genotyping of evolving prokaryotic populations. PeerJ Preprints 4:e2449v1 doi: 10.7287/peerj.preprints.2449v1
for a description. A tutorial is also available. Users need to create a login to use the server. VarCap Galaxy is supported by Department of Microbiology and Ecosystems Science at the University of Vienna.
Semi-Public Galaxy Services
Jetstream was officially launched in September and was added to the Semi-Public Galaxy Services list:
United States: Jetstream
THIS CONTENT HAS BEEN MODIFIED SINCE ITS ORIGINAL PUBLICATION
-
Links:
-
Eligibility:
- Jetstream is part of XSEDE, a "collection of integrated advanced digital resources and services" and is funded by NSF. You need to be eligible for an XSEDE allocation to use Jetstream, which means must be based at a U.S. institution. And although XSEDE is NSF-funded, "projects need not be supported by NSF grants" to receive an allocation.
-
Comments:
- Jetstream enables researchers to launch, use, and shutdown their own Galaxy servers that have been pre-configured similar to the Main Galaxy server. If you want to further customize your launched server you can
become an administratorand evenaccess the server from the shell.
- Jetstream enables researchers to launch, use, and shutdown their own Galaxy servers that have been pre-configured similar to the Main Galaxy server. If you want to further customize your launched server you can
-
User Support:
- See the Galaxy on Jetstream documentation
- See
FAQ
-
Quotas:
- Limits are determined by the size of your allocation and the size of the instances you launch.
-
Sponsor(s):
- The National Science Foundation (NSF)
- Jetstream is based at Indiana University and Jetstream resources are hosted by Indiana University and the Texas Advanced Computing Center (TACC).
Trinity comes to usegalaxy.org
For those looking to perform de novo transcriptome assembly of RNA-seq data, we are pleased to announce that the Trinity assembler is now available in beta on the Galaxy Project's free, public Galaxy server, usegalaxy.org. Due to the high memory requirements of de novo assembly, Trinity runs on the Bridges high memory HPC system at the Pittsburgh Supercomputing Center, via a science gateway grant from XSEDE. Please try the tool out and be sure to report any problems via Galaxy's reporting mechanism.
Galaxy Community Hubs



Share your training resources and experience now Share your experience now
One new trainng resource was added in September:
- Microbial data analysis with Orione, by 鈴木 治夫 (Haruo Suzuki).
Releases
galaxy-lib 16.10.2
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
The September releases work with recents changes to Galaxy and initial mulled scripts and container resolver.
Planemo 0.33.2
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. Planemo saw seven releases in September. The biggest news is that Planemo now uses the brand new Galaxy Tool XML Schema Definition to validate tool definitions. A lot happened with Conda support too.
See GitHub for details.
Earlier Releases
Other packages that have been released in the prior 4 months.
Galaxy v16.07
The Galaxy Committers team is pleased to announce the July 2016 (v16.07) release of Galaxy.
Highlights
Shift of Galaxy tool dependencies to Conda
Galaxy admins obtained the ability to install Galaxy tool’s dependencies using the Conda package manager. This is a Beta feature and we encourage interested deployers to opt-in by modifying configuration. Documentation that explains this switch and answers FAQ is available.
Dynamic tool destinations
Our friends from Canada National Microbiology Laboratory enhanced Galaxy with feature that allows dynamic mapping of tools to destinations based on finer grained admin-specified rules. Please see the wiki. Implemented in PR #2579.
Galaxy chat
Admins can now plug in the included communication server to enable users of their instance to use real-time chat within the Galaxy interface. Please see the documentation to learn how to activate and use this feature. Implemented in PR #2515.
New Galaxy repository
$ git clone -b release_16.07 https://github.com/galaxyproject/galaxy.gitUpdate of existing Galaxy repository
$ git checkout release_16.07 && git pull --ff-only origin release_16.07See our wiki for additional details regarding the source code locations.
See the full release notes for more.
Galaxy Docker Image 16.07
And, thanks to Björn Grüning, there is also now a Docker image for Galaxy 16.07 as well.
CloudBridge 0.1.1
CloudBridge provides a simple layer of abstraction over different cloud providers, reducing or eliminating the need to write conditional code for each cloud. The latest release adds support for advanced networking management and OpenStack Keystone v3 support (required to use NSF Jetstream cloud).
BioBlend 0.8.0
BioBlend is a Python library for interacting with CloudMan and Galaxy‘s API. BioBlend makes it possible to script and automate the process of cloud infrastructure provisioning and scaling via CloudMan, and running of analyses via Galaxy.
See the release notes for what's new in release 0.8.0.
Pulsar 0.7.1 - 0.7.2
Pulsar updates were released in August. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
blend4php 0.1 alpha
The alpha release of the blend4php package, a PHP wrapper for the Galaxy API was announced in June. It follows the lead of BioBlend which provides a Python package for interacting with Galaxy and CloudMan--hence the use of 'blend' in the name of this package. blend4php currently offers a partial implementation of the Galaxy API and includes support for datasets, data types, folder contents, folders, genomes, group roles, groups, group users, histories, history contents, jobs, libraries, library contents, requests, roles, search, tools, toolshed repositories, users, visualizations and workflows.
The motivation for development of this library is for integration with Tripal, an open-source toolkit for creation of online genomic, genetic and biological databases. Integration of Tripal and Galaxy will allow the community research databases to provide next-generation analytical tools to their users using Galaxy. However, this library was created independently of Tripal to support integration of any PHP application with Galaxy.
Please see the API documentation page for full information.
CloudMan 16.04
An update to Galaxy CloudMan on AWS was released in May. CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration or imposed quotas. Once running, you have complete control over Galaxy, including the ability to install new tools.
This is a minor update release with the following changes:
- Galaxy 16.04 update
- Availability on Amazon's Ireland region
- A couple of bug fixes
See the CHANGELOG for a more complete set of changes.
And the rest ...
Other Galaxy packages that haven't had a release in the past four months can be found on GitHub.
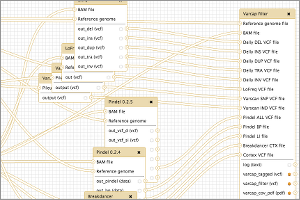
ToolShed Contributions
Tool Shed contributions for September.
Other News
From Ravi Madduri
Globus Genomics RO1 was funded by NHGRI.From Björn Grüning HTTPS support was added to the Galaxy Docker image. Joint work by Marius van den Beek and Alexander Lenail
-