Galaxy Erasmus MC
The homepage of the Erasmus MC Galaxy community
Galaxy is an open-source platform for FAIR data analysis that enables users to:
- use tools from various domains (that can be plugged into workflows) through its graphical web interface.
- run code in interactive environments (RStudio, Jupyter...) along with other tools or workflows.
- manage data by sharing and publishing results, workflows, and visualizations.
- ensure reproducibility by capturing the necessary information to repeat and understand data analyses.
The Galaxy Community is actively involved in helping the ecosystem improve and sharing scientific discoveries.
News
Ten Common Misconceptions About Galaxy (And Why They're Wrong!)
We are thrilled to announce that 'Ten Common Misconceptions About Galaxy (And Why They Are Wrong!)' has just been published in PLOS Computational Biology! This paper is the result of passionate discussions, collaborative debates, and a shared commitment to clarifying what Galaxy truly is—and what it can do. Whether you are a longtime Galaxy user or new to the platform, this paper will challenge assumptions and highlight Galaxy's versatility, scalability, and impact across disciplines.
Bringing Climate, Ecology, and Earth System Science Together
Join Our Galaxy Earth-Climate-Ecology Room
Weekly summary of activity across 150+ galaxyproject repositories
Weekly summary of activity across 150+ galaxyproject repositories
Weekly summary of activity across 150+ galaxyproject repositories
Events
Mar 5 - Mar 6Galaxy Metagenome Training Course 2026
Join us online in the 2026 Metagenome Training Course
Mar 9 - Mar 13Workshop on high-throughput sequencing data analysis with Galaxy
This course introduces scientists to the data analysis platform Galaxy
May 4 - May 8European Geosciences Union (EGU) 2026
A Data Terra session to the European Geoscience Union
FTP Upload
For large files, FTP upload is also available on this server. Connect to port 23 with the same credentials you use to log into Galaxy. Once uploaded, open the Galaxy file upload menu, and click on the Choose FTP file button to import the files into your history.
As an example, a screenshot of the configuration settings for FileZilla can be found here.
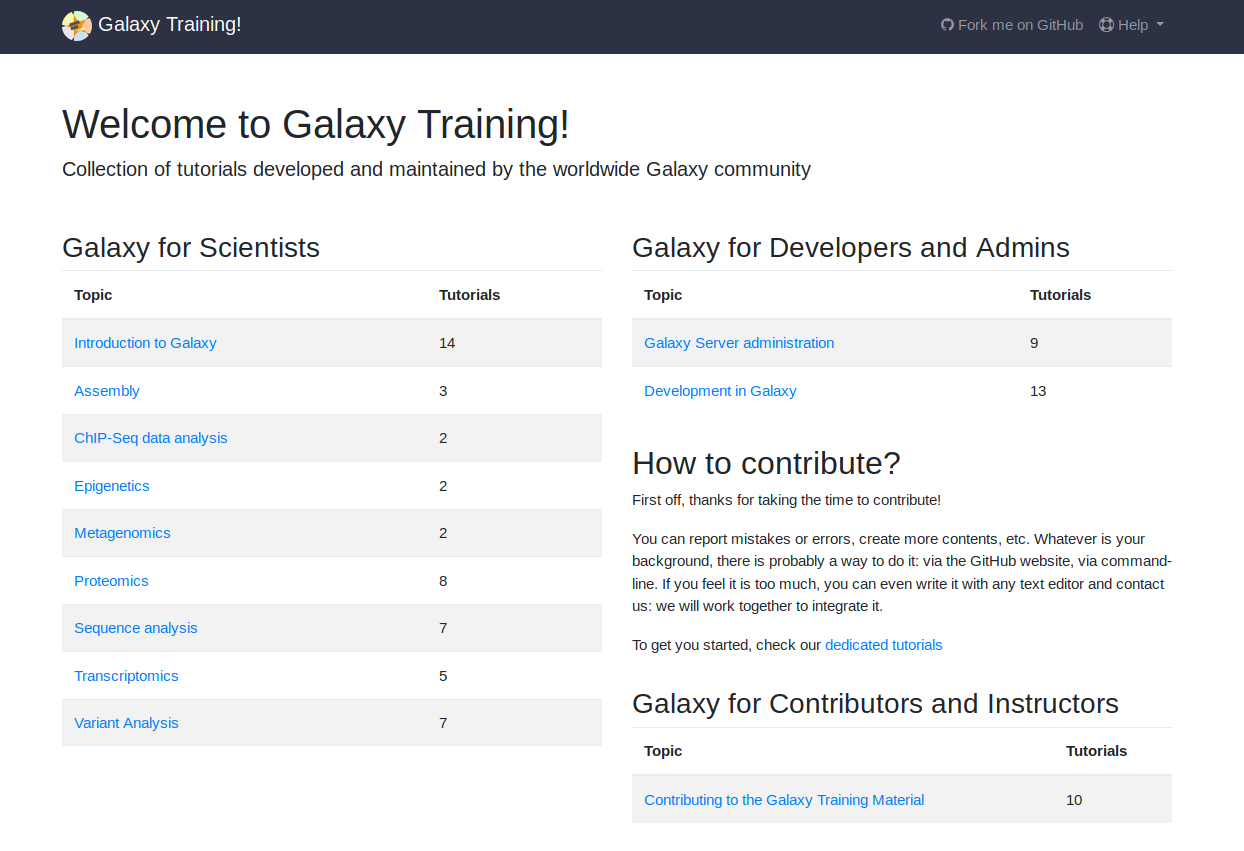
Training
We regularly provide workshops.
But we cannot always meet capacity, so we've put all of our training materials online. This has become a community project with people from all over the world contributing training materials.
Topics include: variant analysis, transcriptomics, metagenomics, epigenetics, and many more!
Our team
This Galaxy is maintained by the Bioinformatics group of the department of Pathology.
For any questions regarding this Galaxy server, please contact us.