Development News Brief
Get Galaxy
new: $ hg clone http://www.bx.psu.edu/hg/galaxy galaxy-dist
upgrade: $ hg pull -u -r e6444e7a1685
# BLAST+ Migration
The tool set NCBI BLAST+ has moved from the Galaxy distribution to the Galaxy Main Tool Shed.
Migration scripts will run upon Galaxy's first launch (after updating to this release) that will automatically handle installing BLAST (and blastxml) from the Tool Shed.
# Reference Genome rsync Server
If you would like to obtain the same reference genome builds and indexes as available on the public Galaxy Main instance, these can retrieved from the rsync server at:
datacache.g2.bx.psu.edu
For example, to download the complete directory for the 'phiX' genome:
$ rsync -avzP rsync://datacache.g2.bx.psu.edu/indexes/phiX .
Genomes are organized in directories by the dbkey. If you are not sure of the dbkey, check your datasets. The dbkey is what is populated into the "database" attribute for a dataset. Read more about how this fits into data integration or [setting up native genome indexes](/admin/NGS Local Setup/).
# More Updates to Output and Error Handling
As reported in the July 20th, 2012 News Brief, several changes have been made to the underlying code that determines run result state from tool exit codes and output. There are now additional enhancements to applying regular expressions and exit code checks. Read more...
# Tools
Admin/Config/Tool Dependencies
-
Enhancements
-
Tophat2 wrapper enhancements:
- Include fusions output. Read more about what this is in the Tophat2 Manual's section Fusion mapping options:
-
Bowtie2 wrapper enhancements:
- Output sorted BAM from Bowtie2 by default
- One benefit is that BAM results can be used as input to Cufflinks without an intermediate sorting step.
- NOTE: If you are using an older version of Bowtie or uploading your own results, sorting is still required before running Cufflinks, whether in SAM or BAM format.
-
-
New
-
Galaxy RNA-seq Analysis Exercise on Main
- Walks through sample protocol step-by-step using paired-end data, initial read QC through CuffDiff analysis
- Includes iGenomes sourced reference annotation GTF, an answer key, and bonus resources
-
# User Interface (UI)
-
Enhancements
- Does not use
enable_tracksorenable_pagesoptions anymore; visualizations and pages are enabled for all instances.
- Does not use
-
New
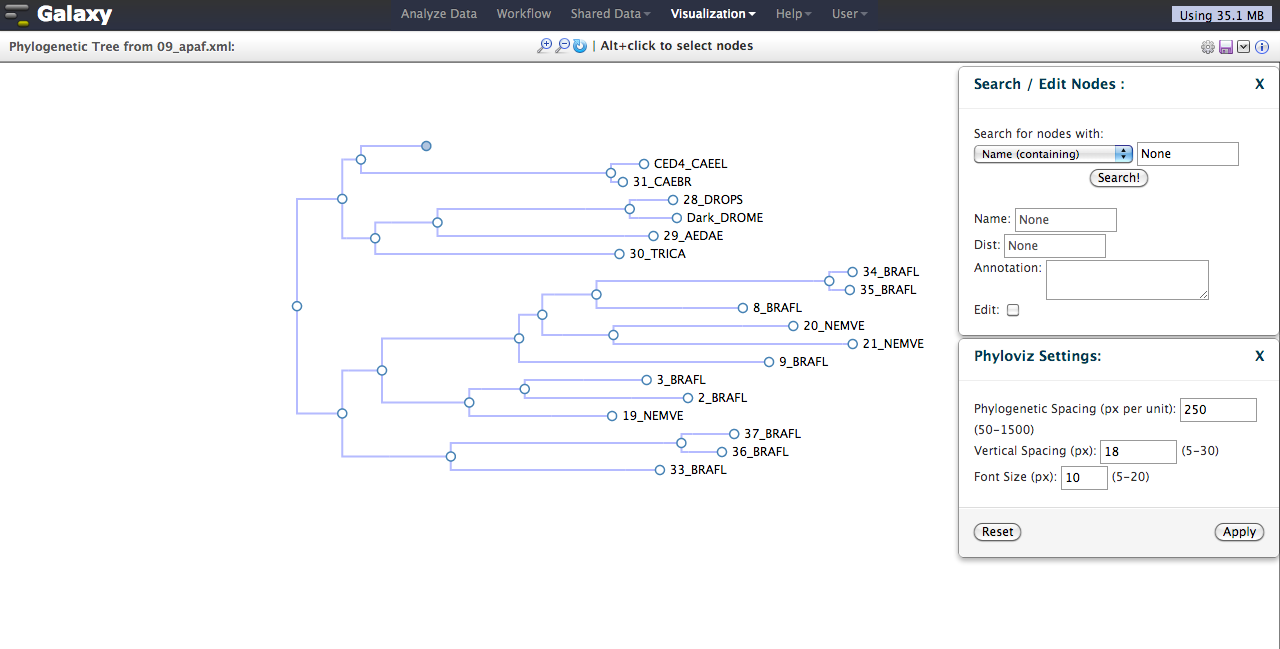
- Addition of interactive phylogenetic tree visualization.