Metagenomics
| Public server: | Server |
|---|---|
| Scope: | Domain specific |
| Summary: | a webserver to process, analyse and visualize Metagenomic and Microbiota data in general. |
comments
More than 200 tools are integrated in this custom Galaxy instance. They were chosen for their use in exploitation of microbiota data.
- The public server is hosted by the UseGalaxy.eu team.
user support
quotas
Storage and computational quotas.
citations
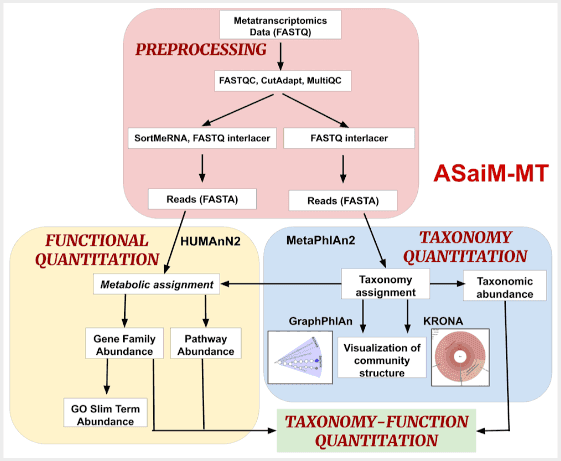
Mehta, S., Crane, M., Leith, E., Batut, B., Hiltemann, S., Arntzen, M. Ø., Kunath, B. J., Delogu, F., Sajulga, R., Kumar, P., Johnson, J. E., Griffin, T. J., & Jagtap, P. D. (2021). ASaiM-MT: A validated and optimized ASaiM workflow for metatranscriptomics analysis within Galaxy framework. F1000Research, 10, 103. doi: 10.12688/f1000research.28608.1
- Batut, B., Gravouil, K., Defois, C., Hiltemann, S., Brugère, J.-F., Peyretaillade, E., & Peyret, P. (2018). ASaiM: A Galaxy-based framework to analyze microbiota data. GigaScience, 7(6). doi: 10.1093/gigascience/giy057
- Metagenomics EU tagged publications in the Galaxy Publication library
sponsors
The Freiburg Galaxy Team but also collectively by groups and individuals from across Europe