June 2017 Galaxy News
Welcome to the June 2017 Galactic News, a summary of what is going on in the Galaxy community. If you have anything to add to next month's newsletter, then please send it to outreach@galaxyproject.org.
GCC2017 is almost here!
GCC2017 will be in Montpellier, France, 26-30 June and will feature two days of presentations, discussions, poster sessions, lightning talks, computer demos, keynotes, and birds-of-a-feather meetups, all about data-intensive biology and the tools and infrastructure that support it. GCC2017 also includes data and coding hacks, and two days of training covering 16 different topics.
GCC2017 will be held at Le Corum Conference Centre in the heart of Montpellier, just 10km from the Mediterranean. This event will gather several hundred researchers addressing diverse questions and facing common challenges in data intensive life science research. GCC participants work across the tree of life, come from around the world, and work at universities, research organizations, industry, medical schools and research hospitals.
Regular Registration Rates Extended to June 16
If you missed the May 31 regular registration deadline then don't worry because it has been extended to June 16. Regular registration starts at 75€ / day for post-docs and students. You can also book low cost conference housing when you register.
Keynotes
Eytan Domany
Personalized Analysis of Cancer Data: From Genes to Pathways (and Back)
Dr. Eytan Domany will cover his work in human genomics and computational biology principally in the cancer domain. Eytan Domany is a Professor of Computational Systems Biology in the Department of Physics and Complex Systems at the Weizmann Institute of Science in Rehovot, Israel. The main focus of his group is to mine data from large-scale experiments in biology. The work includes development of mathematical methods, their implementation in algorithms (which are incorporated in user-friendly computational tools), which are then applied to study biological data. Eytan was awarded the Sergio Lombroso Award in Cancer Research in 2016 for his research efforts.
Anton Nekrutenko & James Taylor
The Galaxy Project: from the Planck epoch to Montpellier
Dr. Anton Nekrutenko and Dr. James Taylor will present a keynote reviewing the history of the Galaxy project, and how it went from a humble Perl script to having its own international meeting in the South of France in the summer of 2017.
If you are reading this, you probably already know who they are. If not, see their bios here.
And the rest of the schedule ...
... will be published this week on the conference schedule web site.
Submit a Lightning Talk!
Lightning talks are an excellent chance to share your work, preliminary or otherwise, with the community. Lightning talks are 7 minutes long and are the quickest way to make a lasting impression on, and get feedback from all participants. Submit your lightning talk proposal by June 23.
Talk, Poster and Demo Submission is Closed, but ...
Abstract submission for regualr talks, posters and demos closed in May. However you can still submit late oral prentations abstracts, posters and demos. Late oral presentation abstracts are considered as cancellations occur, and posters and demos will be reviewed until the space is full.
GCC2017 Sponsors
GCC2017 has 11 sponsors! Sponsors are potential partners in your research and sponsorships greatly lower the cost of conference registration. Have a look at the GCC2017 sponsor list and plan on learning what they have to offer (and thanking them) while you are in Monpellier.
Hack the Universe: Capture the Flag at GCC2017!
GCC2017 Call for BoFs!
There is no better place than a Galaxy Community Conference to meet and learn from others doing data-intensive biology. GCC2017 continues this tradition by again including Birds of a Feather (BoF) meetups. Birds of a Feather meetups are informal gatherings where participants group together based on common interests.
Interested?
Galaxy Tutorial @ ISMB/ECCB 2017
Know someone who needs to learn more about deploying Galaxy in their organization? There will be a half day Making Galaxy Work for You tutorial at ISMB/ECCB 2017 in Prague, Czechia. The tutorial will provide a practical, hands-on guide to adapting the Galaxy platform to the specific needs of individuals attending the ISMB.
Participants will learn to
- Create Galaxy compatible tool and workflow definitions that are publicly accessible and that make it easy for any instance administrator to add your work to their server.
- Deploy Galaxy and scale it up to target production-ready resources such as a Postgres database, NGINX webserver, and distributed resource managers such SLURM or PBS.
Interested? Sign up when you register for ISMB/ECCB (and register before the tutorial fills up).
We also expect a lot of other Galaxy-related content at ISMB/ECCB 2017 and BOSC 2017. We'll add the presentations to the Galaxy @ ISMB/ECCB 2017 & BOSC 2017 event page as the schedules are posted. (And, please let us know if you have anything on the program.)
See you in Prague!
All events
There are a plenitude of Galaxy related events coming up in the next few months:
See the Galaxy Events Google Calendar for details on other events of interest to the community.
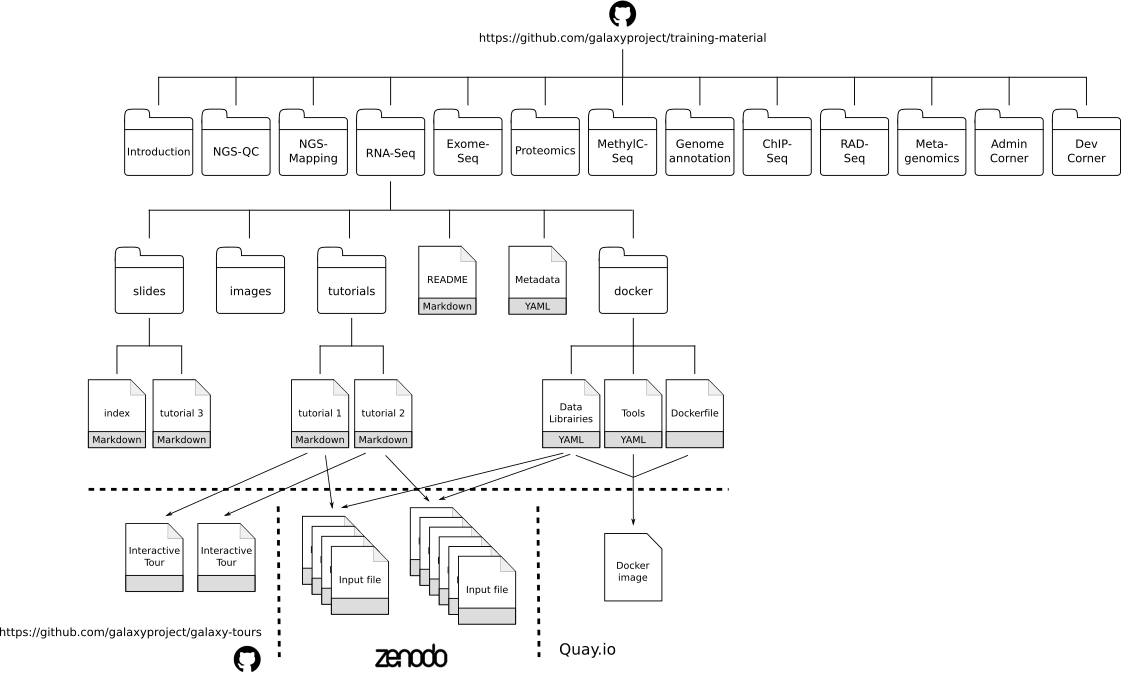
Training Material Re-use Hackathon Reports
The ELIXIR/GOBLET/GTN hackathon for Galaxy training material re-use was held last month in Cambridge, UK. A lot was accomplished:
New Publications
85 new publications referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in May.
Some highlights from the papers added last month:
- GSuite HyperBrowser: integrative analysis of dataset collections across the genome and epigenome Boris Simovski, Daniel Vodák, Sveinung Gundersen, Diana Domanska Abdulrahman Azab, Lars Holden, Marit Holden, Ivar Grytten, Knut Rand, Finn Drabløs Morten Johansen, Antonio Mora, Christin Lund-Andersen, Bastian Fromm Ragnhild Eskeland, Odd Stokke Gabrielsen, Egil Ferkingstad, Sigve Nakken Mads Bengtsen, Alexander Johan Nederbragt, Hildur Sif Thorarensen Johannes Andreas Akse, Ingrid Glad, Eivind Hovig, Geir Kjetil Sandve. GigaScience (27 April 2017), doi:10.1093/gigascience/gix032
- Coherent Application Delivery on Hybrid Distributed Computing Infrastructures of Virtual Machines and Docker Containers German Molto, Miguel Caballer, Alfonso Perez, Carlos Alfonso, Ignacio Blanquer. In 2017 25th Euromicro International Conference on Parallel, Distributed and Network-based Processing (PDP) (2017), pp. 486-490, doi:10.1109/pdp.2017.29
- ChimeRScope: a novel alignment-free algorithm for fusion transcript prediction using paired-end RNA-Seq data You Li, Tayla B. Heavican, Neetha N. Vellichirammal, Javeed Iqbal, Chittibabu Guda Nucleic Acids Research (02 May 2017), doi:10.1093/nar/gkx315
- Jupyter and Galaxy: Easing entry barriers into complex data analyses for biomedical researchers Björn A. Grüning, Helena Rasche, Boris Rebolledo-Jaramillo, Carl Eberhard, Torsten Houwaart, John Chilton, Nate Coraor, Rolf Backofen, James Taylor, Anton Nekrutenko. PLOS Computational Biology, Vol. 13, No. 5. (25 May 2017), e1005425, doi:10.1371/journal.pcbi.1005425
- MAGenTA: a Galaxy implemented tool for complete Tn-Seq analysis and data visualization Katherine M. McCoy, Margaret L. Antonio, Tim van Opijnen. Bioinformatics (11 May 2017), doi:10.1093/bioinformatics/btx320
Publication Topics
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 49 | methods | 21 | usepublic | 16 | usemain | 15 | workbench | |||
| 10 | refpublic | 5 | tools | 5 | isgalaxy | 4 | cloud | |||
| 3 | reproducibility | 2 | uselocal | 1 | shared | 1 | project | |||
| 1 | unknown | 1 | other |
Who's Hiring

The Galaxy is expanding! Please help it grow.
- Bioinformatics Research Associate, Huttenhower Lab, Harvard School of Public Health, Boston, Massachusetts, United States
- Adviser for Data Infrastructure, SURFsara, Amsterdam, The Netherlands
- Automated deployment of Galaxy environments using Ansible, Institut de Biologie Paris Seine, France
- Scientific Research Programmer, Sethuraman Lab, California State University San Marcos. Develop model-based population genomics pipelines.
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
Public Galaxy Server News
There are over 90 publicly accessible Galaxy servers and six semi-public Galaxy services. Here's what happened with them in May
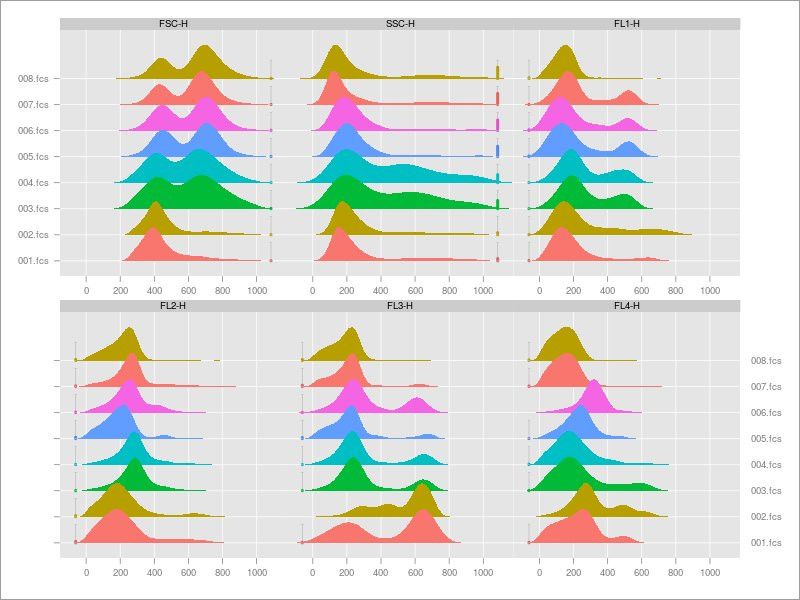
ImmPort Galaxy Update
A new release of ImmPort Galaxy is out:
- R is all up to date logicle trans,
- Added a transformation algorithm to the Automated gating
- Transformation and Converson of FCS files to text tool,
- And a couple of tools to generate 1D Density plots.
More cool visualizations to look at your flow cytometry data!
Usegalaxy.org
- The STAR RNA-seq ultra fast aligner is now on Usegalaxy.org and has 64GB of memory available for each job.
- TopHat is gone from usegalaxy.org because there are now superior alternatives.
New Public Galaxy Servers
GVL MEL
Galaxy Melbourne is a general purpose Galaxy based on the Genomics Virtual Lab platform. Support options include GVL Help, tutorials at GVL Learn and Galaxy Tut. Registration is required but anyone can register. The default quota of 100GB which can be increased on request. GVL MEL is supported by the Genomics Virtual Lab, Melbourne Bioinformatics, and Nectar.
ChimeRScope
ChimeRScope Galaxy is a tool publishing server for fusion gene discovery from transcriptome-sequencing (RNA-seq) datasets. ChimeRScope is a java application that uses a novel alignment-free approach for discovering fusion genes from transcriptome-sequencing (RNA-seq) datasets. Prediction results from simulated datasets and real datasets show that our method achieves the best prediction accuracy, comparing to all other popular methods.
See You Li, Tayla B. Heavican, Neetha N. Vellichirammal, Javeed Iqbal, Chittibabu Guda; ChimeRScope: a novel alignment-free algorithm for fusion transcript prediction using paired-end RNA-Seq data. Nucleic Acids Research 2017 gkx315. doi: 10.1093/nar/gkx315
There is a ChimeRScope Manual including a FAQ. The server supports anonymous access and creation of user logins. ChimeRScope Galaxy is supported by the Guda Lab at the University of Nebraska Medical Center.
Tools
Qiime and mothur
Galaxy has reached many different communities far beyond classical NGS. To support this the IUC is organising regularly contribution fests dedicated to special topics, with the aim to increase tool coverage and develop re-usable workflows. One of the first contribution fests focused on metagenomics, a field that is gaining more traction in the Galaxy community. Here we highlight two pull requests that were recently merged and added Qiime (Quantitative Insights Into Microbial Ecology) and mothur to Galaxy. These tools are the most popular pipelines used by the microbial ecology community to perform microbiome analyses from raw DNA sequencing data.
Based on previous work by James Johnson, ~150 tools were developed and adapted to the most recent IUC standards for Galaxy tools, including integration with BioConda and BioContainers. This effort was led by Saskia Hiltemann and Bérénice Batut and several reviewers and contributors, who created more than 300 commits and touched more than 600 files (including test-data). Thanks Saskia and Bérénice for this effort!
The tools can be installed via the Galaxy Tool Shed:
Both tool suites and many more metagenomics tools are integrated into the metagenomic Galaxy flavor.
ToolShed Contributions
Tool Shed contributions from May.
Releases
May 2017 Galaxy Release (v 17.05)
The Galaxy Committers published the 17.05 release of Galaxy in May.
Tag your data with propagating hashtags
- Large Galaxy histories used to be messy. Hashtags make it easy to track dataset (and collection) relationships.
- These two movies (both under a minute) explain how to use this with datasets and with collections.
- Learn more about Galaxy histories in our updated tutorial.
Drag & Drop datasets into tool inputs
- Interface now allows dragging datasets from history panel into the content selectors of the tool form.
- Implemented in Pull Request 3871.
Upload directly to a collection
- You can now bypass the history manipulation and upload your data straight into a collection for convenience.
- Learn more about collections and how to use them in a new tutorial.
We extend special thanks to the 64 New Contributors to Galaxy in the past year.
Additionaly there are New Configuration Options and New Datatypes sections.
Galaxy Docker Image 17.05
The Galaxy Docker project has seen a new release, following Galaxy 17.05. Major features are an additional Docker compose setup with SLURM and HT-Condor deployments, BioContainers integration and much more automatic testing.
And
- The Galaxy Docker Project has reached more than 23k downloads on Dockerhub - not counting quay.io and all flavors
-
From John Chilton:
- Awesome new mode for docker-galaxy-stable by Björn Grüning: container scheduling with Condor
Parsec 1.0.0 - 1.0.2
Parsec is a newly released set of command-line utilities to assist in working with Galaxy servers. It uses automatically generated wrappers for BioBlend functions. Manage histories, launch workflows, and more, all from the command line. The README includes several examples.
Planemo 0.41.0
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. These releases included numerous fixes and enhancements.
See GitHub for details.
BioBlend 0.9.0
BioBlend is a Python library for interacting with CloudMan and Galaxy‘s API. BioBlend makes it possible to script and automate the process of cloud infrastructure provisioning and scaling via CloudMan, and running of analyses via Galaxy.
See the release notes for what's new in release 0.9.0.
galaxy-lib 17.5.9-11
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
Galaxy CloudMan 17.05 on AWS
A new release of Galaxy CloudMan is available on the Amazon Web Services cloud infrastructure. This release includes Galaxy 17.05, an updated tool list, and Slurm configuration changes to improve job performance. To get started, vist https://beta.launch.usegalaxy.org/ or take a look at the Getting Started guide.
Earlier Releases
Other packages that have been released in the prior 4 months.
CloudLaunch
Technically, the all-new Galaxy CloudLaunch service has been in public beta since February but keep in mind that it will replace the current CloudLaunch service eventually so give it a try and let us know how it performs for you.
CloudBridge 0.2.0
CloudBridge aims to provide a simple layer of abstraction over different cloud providers, reducing or eliminating the need to write conditional code for each cloud. It is currently under development and is in an Alpha state. Release 0.2.0 includes several fixes and enhancements.
And the rest ...
Other Galaxy packages that haven't had a release in the past four months can be found on GitHub.
Other News
-
From Paul Gordon
-
From Björn Grüning
- There is a new HiCExplorer Galaxy Docker version, based on Galaxy 17.05, made possible by Fidel Ramírez and friends.
- From Mo Heydarian
- Creating Galaxy workflows is easier than you think. If you like reproducibility, you'll like workflows!
- New video on Creating collections by uploading from SRA.
-
From Nate Coraor
- Heads up: Galaxy roles published on galaxy.ansible.com have changed namespaces from
galaxyprojectdotorgto justgalaxyproject
- Heads up: Galaxy roles published on galaxy.ansible.com have changed namespaces from