Galactic November News!
Welcome to the November 2015 Galactic News, a summary of what is going on in the Galaxy community.
If you have anything to include in the next News, please send it to [Galaxy Outreach](mailto:outreach AT galaxyproject DOT org).
New Papers
81 new papers referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in October. Highlights include:
- Genomics Virtual Laboratory: A Practical Bioinformatics Workbench for the Cloud
Enis Afgan, Clare Sloggett, Nuwan Goonasekera, Igor Makunin, Derek Benson, Mark Crowe, Simon Gladman, Yousef Kowsar, Michael Pheasant, Ron Horst, Andrew Lonie, PLoS ONE October 26, 2015 DOI: 10.1371/journal.pone.0140829.
See also Australian-made Genomics Virtual Laboratory: A Practical Bioinformatics Workbench for the Cloud, .... - Analyzing HT-SELEX data with the Galaxy Project tools - a web based bioinformatics platform for biomedical research William H. Thiel, Paloma H. Giangrande, Methods (October 2015), doi:10.1016/j.ymeth.2015.10.008
- ReproPhylo: An Environment for Reproducible Phylogenomics
Amir Szitenberg, Max John, Mark L. Blaxter, David H. Lunt, PLoS Comput Biol, Vol. 11, No. 9. (3 September 2015), e1004447, doi:10.1371/journal.pcbi.1004447 - RNA-Seq UD: A bioinformatics platform for RNA-Seq analysis Miguel Ramirez, Cristian A. Rojas-Quintero, Nelson E. Vera-Parra, in "Information Systems and Technologies (CISTI), 2015 10th Iberian Conference on" (June 2015), pp. 1-5, doi:10.1109/cisti.2015.7170565
- BioImg.org: A Catalog of Virtual Machine Images for the Life Sciences Martin Dahlö, Frédéric Haziza, Aleksi Kallio, Eija Korpelainen, Erik Bongcam-Rudloff and Ola Spjuth, Bioinformatics and Biology Insights 2015:9 125–128 doi: 10.4137/BBI.S28636
- SBMLsqueezer 2: context-sensitive creation of kinetic equations in biochemical networks Andreas Dräger, Daniel C. Zielinski, Roland Keller, et al., BMC Systems Biology, Vol. 9, No. 1. (09 October 2015), 68, doi:10.1186/s12918-015-0212-9
- RDF2Graph a tool to recover, understand and validate the ontology of an RDF resource Jesse C. J. van Dam, Jasper J. Koehorst, Peter J. Schaap, Vitor A. P. Martins dos Santos, Maria Suarez-Diez, Journal of Biomedical Semantics, Vol. 6, No. 1. (23 October 2015), 39, doi:10.1186/s13326-015-0038-9
- MicRhoDE: a curated database for the analysis of microbial rhodopsin diversity and evolution Dominique Boeuf, Stéphane Audic, Loraine Brillet-Guéguen, Christophe Caron, Christian Jeanthon, Database, Vol. 2015 (01 January 2015), bav080, doi:10.1093/database/bav080
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | Cloud | - | Project | 5 | Tools | 18 | UsePublic | |||
| 2 | HowTo | 7 | RefPublic | - | UseCloud | 1 | Visualization | |||
| 5 | IsGalaxy | 3 | Reproducibility | 6 | UseLocal | 16 | Workbench | |||
| 43 | Methods | - | Shared | 8 | UseMain |
Events
GCC2016 Training Topic Vote Ends This Friday
Thanks to everyone who nominated topics for GCC2016 Training. Please take a few minutes to vote on the topics you want to see offered at GCC2016 Training.
The 2016 Galaxy Community Conference (GCC2016) features two full days of training on June 26-27. Each day will have multiple tracks, with each track featuring several sessions throughout the day. GCC2016 will be held at Indiana University in Bloomington, Indiana, United States.
Topic voting closes November 6.
Topics will then be selected and scheduled based on topic interest, and the organizers' ability to confirm instructors for each session. Some very popular sessions may be scheduled more than once. The final schedule will be posted before registration opens (which we are hoping will be the end of November).
IUC Contribution Fest - Metagenomics Tools and Workflows
We are planning a remote hackathon on 30th of November and 1st of December for developers to work mainly on Galaxy metagenomic tools.
The remote nature of this gives people who don't have the opportunity to come to GCC hackathons (which have always been productive and a lot of fun) a chance to participate in a Galaxy hackathon. Having a well defined topic will allow us to accomplish a lot and let people who don't have particular tasks in mind find something to work on very quickly.
We are collecting ideas to work on, but we expect to attract the most participation by simply getting tool developers interested in getting help adding collection support to their existing tools and workflows to show up and participate.
If you are interested in participating in the hackathon but not interested in actual tool development - we will assemble a list of smaller, manageable Python and JavaScript tasks to work on and certainly documentation is a chronically lacking for collections so we could use help there and no actual coding would be required.
We encourage ideas or advice about how to organize this so please let us know. A core group will be available on IRC all day and we will have 4 google hangouts across those days to organize, answer questions, and report progress.
We will do our best to coordinate and make this hackathon a nice and productive experience and we would like to especially focus on working reasonable hours and discourage overnighters.
All forms of contribution are welcome!
See the event home page for more.
Call for GCC2017 Hosts
      |
|||||
| GCC2017 needs a host! (and a logo) |
|||||
The call for hosts for GCC2017 closes 30 November. If you are interested, please see this announcement.
October GalaxyAdmins Meetup Video and Slides
Video and slides for the GalaxyAdmins meetup on October 15 are now available. David Kovalic presented on Galaxy at Analome – Outreach, Opportunity and Challenges. This was followed by an active discussion about plans to migrate this wiki to another platform.
GalaxyAdmins is a special interest group for Galaxy community members who are responsible for Galaxy installations. We're skipping the December meetup (wouldn't you rather be on holiday?) so the next meetup will be in February. Hope to see you there.
Upcoming Events
There are upcoming events on 3 continents in 4 countries, and a veritable globular cluster of events in France this month. See the Galaxy Events Google Calendar for details on other events of interest to the community.
| |
Designates a training event offered by GTN member(s) |
Who's Hiring
The Galaxy is expanding! Please help it grow.
- Postdoc developing advanced MS-based metabolomics bioinformatics methods, University of Minnesota. Closes November 30.
- Software Developer / Bioinformatician, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany
- The Galaxy Project is hiring software engineers and post-docs
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
New Public Galaxy Servers
1 new publicly accessible Galaxy servers was added in October, bringing the total dangerously close to 80.
Galaxy@PRABI
-
Links:
-
Domain/Purpose:
- A general purpose Galaxy instance that includes bioinformatics tools developed by the research teams working in the perimeter of the PRABI core facility, including kissplice/kissDE, TETools, SexDetector, and priam available through our local toolshed instance.
-
Comments:
- Dedicated server: PowerEdge R920 Rack Server - 64 CPU, 512 GB RAM, 15To GB of disk space.
-
User Support:
- galaxy@prabi user list (in French)
- Email support: [galaxy DASH support AT listes DOT univ-lyon1 DOT fr](mailto:galaxy DASH support AT listes DOT univ-lyon1 DOT fr)
- We also provide galaxy training courses for RNA-seq and ChIP-seq data analysis.
-
Quotas:
- Disk space is limited to 100 MB for unregistered users, registered users are free to use up to 50 GB.
- Users from the FRBioenvis and LECA teams are free to use up to 250 GB disk space.
- Can be increased up to 1-2 TB in collaborative projects (please contact [galaxy@PRABI Admins](mailto:galaxy DASH support AT listes DOT univ-lyon1 DOT fr).)
-
Sponsor(s):
RDF2Graph
-
Link:
-
Domain/Purpose:
- RDF2Graph recovers the structure of an RDF resource.
-
Comments:
- Graphs can be exported to Cytoscape, OWL, and ShEx.
- See "RDF2Graph a tool to recover, understand and validate the ontology of an RDF resource" by van Dam et al., Journal of Biomedical Semantics 2015, 6:39 (2013)
-
User Support:
-
Quotas:
- Server can be used by anyone, but you'll need to create an account.
- The RDF resource is limited to 20,000,000 lines.
-
Sponsor(s):
Releases
Planemo 0.15.0 - 0.18.1
Planemo saw a meteor storm (sorry!) of activity in October with 4+ releases.
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. Planemo 0.18.1 is the most recent release. See the release history.
BioBlend 0.7.0
BioBlend version 0.7.0 was released few days ago. BioBlend is a python library for interacting with CloudMan and the Galaxy API. CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration.) From the release CHANGELOG.
Other Releases
July 2015 Galaxy Release (v 15.07)
Galaxy Release v 15.07 includes enhancements to Interactive Environments and the Workflow Editor, an update of the Policies for Committers and Pull Requests and much more.
Mid 2015 Galaxy CloudMan Release
Security Advisories
Three security vulnerabilities were announced and patched in August. The Galaxy Committers have released a number of fixes which have been rolled in to affected versions of Galaxy dating back to the 14.10 release.
Pulsar Pulsar 0.5.0 was released in May. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
**CloudMan ** The most recent edition of CloudMan was released in August.
blend4j v0.1.2 blend4j v0.1.2 was released in December 2014. blend4j is a JVM partial reimplemenation of the Python library bioblend for interacting with Galaxy, CloudMan, and BioCloudCentral.
Galaxy Community Hubs
 |
 |
 |
| Share your training resources and experience now | Share your experience now |
2 new Galaxy Training Network members were added last month:
- Noor Biotechnologies Ltd. Noor Biotechnologies has a presence in Malaysia and Saudi Arabia, and is available for training worldwide.
- University of Minnesota Minnesota Supercomputing Institute Research Informatics Solutions RIS is a group of experienced bioinformatics experts within the Supercomputing Institute at the University of Minnesota.
And 2 new Community Log Board entries were added as well. Both are about wrapping Tools for Galaxy:
- Galaxy Tool Generating Dataset Collections Steve Cassidy documented how create dataset collections in Galaxy tool definitions. (And Alveo is also a cool, non-life science application of Galaxy.)
- How Galaxy resolves dependencies (or not) Peter van Heusden documents tool dependencies that specify how to source the actual packages that implement a tool’s commands.
ToolShed Contributions
Ran out of time to compile these. Look for a double batch in December.
Other News
- Have a look at a fresh chart of Main's usage showing the improvements in Galaxy reports by Daniel Bouchard.
-
From Björn Grüning:
- 💕 this community! DESeq2 update requested by @mikelove: the Galaxy wrapper is available before BioC version
-
From John Chilton:
- Versioned Tap Hackathon Proposal
- Galaxy visualizations from Eteri Sokhoyan and powered by Cytoscape.js have been merged.
-
- A list of tools presented at Genome Informatics 2015 that might be of interest to the Galaxy community.
-
From Oleg Shpynov
-
From Dannon Baker:
- Det'Rprok workflow from this paper updated at MyExperiment.
- We decided not to have a Data Carpentry / Software Carpentry Train the Trainer event before GCC2016. However, we have nominated a topic on Best Practices and Resources for Galaxy Training. If that interests you then please vote for it.
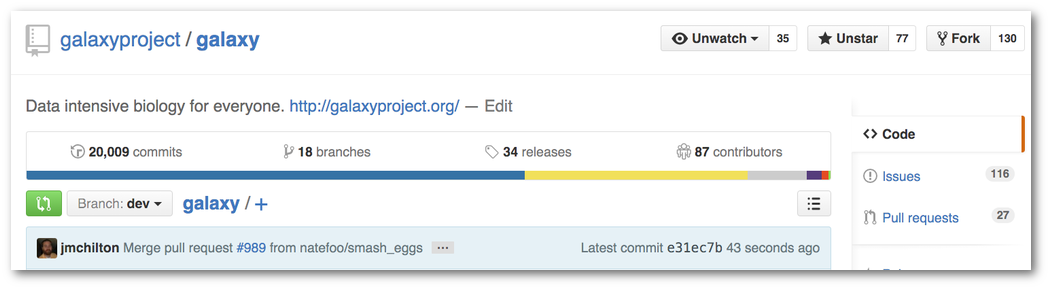
Galaxy Codebase Reaches 20 000 Commits
In September Galaxy turned 10 years old by one important measure. Another milestone was reached in October when the Galaxy codebase surpassed of 20000 commits. And appropriately (and if I can count), the 20,000th was a pull request commit from community member Helena Rasche originally submitted by team member Nate Coraor.
Our thanks go to the community and the team alike.