Galaksio: a Galaxy user interface focused on running prepared workflows
Make your workflows more accessible to non-bioinformaticians
Galaksio (Esperanto for Galaxy) is a user interface for Galaxy designed to make workflows more accessible to non-bioinformaticians. The interface is focused on running prepared workflows and bioinformaticians can use the interface to make comprehensive workflows accessible for colleagues lacking the skills necessary to create their own workflows. The Galaksio interface runs on its own Python web server and can be installed from our Github repository or by using the docker compose file provided in the repository.
Features
The user interface is currently in beta and especially the handling of input parameters in different workflows is under development along with the addition of further features (see table).
| Feature | Category | Implemented | Planned |
|---|---|---|---|
| User sign-in/out | Users | ✅ | |
| User sign-up | Users | ✅ | |
| Workflow listing | Workflows | ✅ | |
| Workflow importing | Workflows | ✅ | |
| Workflow execution | Workflows | ✅ | |
| Workflow creation | Workflows | ✔ | |
| Simultaneous execution of workflows | Workflows | ✅ | |
| Recovering previous executions | Workflows | ✅ | |
| Help and description for tools in workflow | Workflows | ✅ | |
| Input selection and parameter configuration | Workflows | ✅ | |
| History selection | History | ✅ | |
| History creation | History | ✔ | |
| History deletion | History | ✔ | |
| Dataset uploading | Dataset manipulation | ✅ | |
| Dataset downloading | Dataset manipulation | ✅ | |
| Dataset deletion | Dataset manipulation | ✅ | |
| Dataset collection creation | Dataset manipulation | ✅ | |
| Dataset collection deletion | Dataset manipulation | ✔ | |
| Tool execution | Tools | ✔ |
Architecture
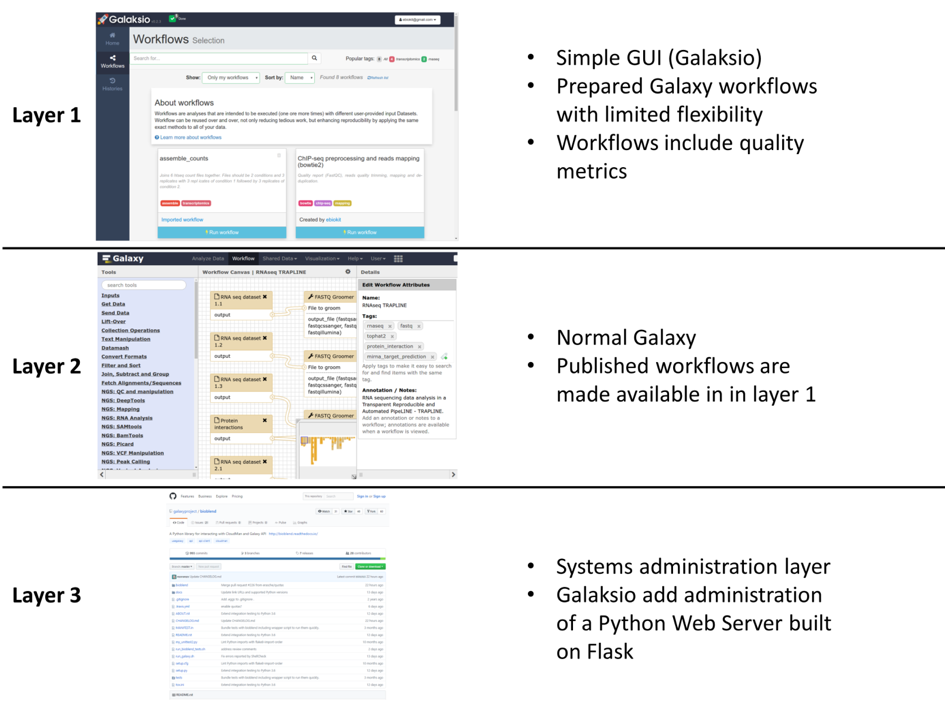
The key contribution of the Galaksio interface is that it provides bioinformaticians with an easy way to make high quality workflows available to researchers with minimal training in bioinformatics. As a part of the B3Africa project (http://www.b3africa.org) we call this “a layered approach” where users of three different skill levels are expected to use the system (see figure).
- Researchers with limited training in bioinformatics are able to access prepared workflows using the Galaksio interface.
- Bioinformaticians can design and develop workflows using the normal Galaxy interface and any tools developed for Galaxy.
- Systems administration is carried out exactly like on a normal Galaxy server. The only addition to maintenance is the small Python webserver that hosts a Galaksio website and forward any commands to the Galaxy server.
A Galaksio interface is always pointing towards a Galaxy server which hosts all the data and executes the workflows. During testing we have operated with our own locally hosted Galaxy server but also tested it using usegalaxy.org and even organised a training workshop with the support of the Freiburg Galaxy Team using the galaxy.uni-freiburg server. The benefit of this set up is that any Galaxy server which provides API access can be provided with a Galaksio interface without any modifications to the server itself. Meaning that Galaksio is easy to add to existing Galaxy server in order to improve access to best practice workflows or complex tool packages.
Figure 1, image from the article "Galaksio, a user friendly workflow-centric front end for Galaxy" (https://doi.org/10.14806/ej.23.0.897).
Further reading
- Github repository: https://github.com/fikipollo
- Software documentation: http://galaksio.readthedocs.io/en/latest/
- The marker paper of the Galaksio interface: https://doi.org/10.14806/ej.23.0.897
- (For real enthusiasts) A thesis describing the need of more efficient division of labour in life science and the role of Galaksio in this: https://pub.epsilon.slu.se/14669/