The Italian Galaxy server
The Italian Galaxy server UseGalaxy.it is maintained primarily by ELIXIR-ITALY, in collaboration with many academic groups across Italy. Please check our Terms of Use before using the server.
The following Institutions are actively providing resource to usegalaxy.it: CNR, INFN, University of Bari, Consortium GARR, CINECA and University of Milan.
Get information
Be updated on the Italian Galaxy community activities! Subscribe to our mailing list.
Our Data Policy
| Registered Users | Unregistered Users | GDPR Compliance |
|---|---|---|
| User data on UseGalaxy.it (i.e. datasets, histories) will be available as long as they are not deleted by the user. Once marked as deleted the datasets will be permanently removed within 14 days. If the user "purges" the dataset in the Galaxy, it will be removed immediately, permanently. In special cases, an extended quota can be requested for a limited time period.. | Processed data will only be accessible during one browser session, using a cookie to identify your data. This cookie is not used for any other purposes (e.g. tracking or analytics). If UseGalaxy.it service is not accessed for 90 days, those datasets will be permanently deleted. | The Galaxy service complies with the EU General Data Protection Regulation (GDPR). You can read more about this on our Terms and Conditions. |
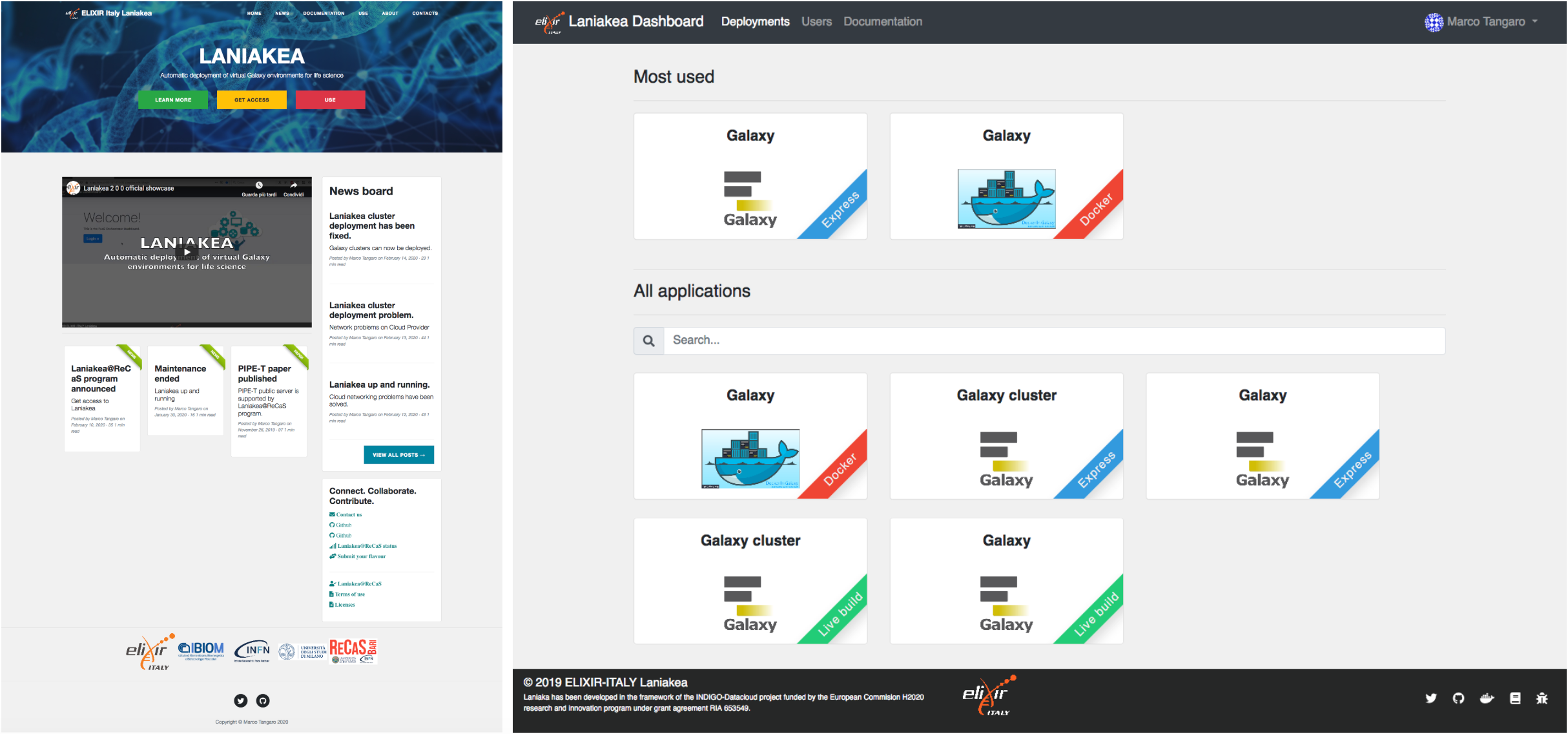
Laniakea@ReCaS
The Laniakea@ReCaS service, based on Laniakea software stack, provides the possibility to automate the creation of Galaxy-based virtualized environments through an easy setup procedure, providing an on-demand workspace ready to be used by life scientists and bioinformaticians. At the end of the configuration process, the user gains access to a private, production-grade, fully customizable, Galaxy virtual instance. Laniakea features the deployment of a stand-alone or cluster-backed Galaxy instances, shared reference data volumes, encrypted data volumes and rapid development of novel Galaxy flavours for specific tasks.
For more information about Laniakea, please subscribe to our mailing list.
Communities: Laniakea's public servers
Vinyl
VINYL (Variant prIoritizatioN bY survivaL analysis) integrates an innovative method for variant prioritization, along with a highly curated collection of databases and resources for the annotation of human genetic variants.
CorGAT
CorGAT, the Coronavirus Genome Analysis Tool, is a novel, highly effective and user-friendly approach for the functional annotation of SARS-CoV-2 genomes based on Galaxy.
ITSoneWB
ITSoneWB (ITSone WorkBench) collects and integrates the high-quality ITS1 reference collection in ITSoneDB with DNA metabarcoding well-established analysis pipelines and new tools in an easy-to-use service addressing the eukaryotic domain of life.
PIPE-T
PIPE-T is the first Galaxy tool for parsing, filtering, normalising, imputing, and analysing RT-qPCR data. PIPE-T integrates the functionalities implemented in various R packages, such as HTqPCR, impute, and RankProd, in a simple, transparent, accessible, reproducible, and user-friendly environment.